DifferentialNet
DifferentialNet is a novel database that provides users with differential interactome analysis of human tissues (https://netbio.bgu.ac.il/diffnet/). Users query DifferentialNet by protein, and retrieve its differential protein–protein interactions (PPIs) per tissue via an interactive graphical interface. To compute differen- tial PPIs, we integrated available data of experimen- tally detected PPIs with RNA-sequencing profiles of tens of […] More Info | Go to DifferentialNet

MotifNet
Network motifs are small topological patterns that recur in a network significantly more often than expected by chance. Their identification emerged as a powerful approach for uncovering the design principles underlying complex networks. However, available tools for network motif analysis typically require download and execution of computationally intensive software on a local computer. We present […] More Info | Go to MotifNet
MyProteinNet
The identification of the molecular pathways active in specific contexts, such as disease states or drug responses, often requires an extensive view of the potential interactions between a subset of proteins. This view is not easily obtained: it requires the in- tegration of context-specific protein list or expres- sion data with up-to-date data of protein […] More Info | Go to MyProteinNet
ODiseA
The Organ-Disease Annotation (ODiseA) database of tissue-selective hereditary diseases is a manually-curated resource of hereditary diseases and their inflicted tissues. Accompanies the paper Hekselman et al, JMB 2022.
More Info | Go to ODiseA
ProAct
The Process Activity (ProAct) webserver estimates the preferential activity of biological processes in tissues, cells, and other contexts. It can also be used to annotate cell subsets inferred from single-cell transcriptomics. Accompanies the paper Sharon et al, Nucleic Acids Research 2023.
More Info | Go to ProAct
ResponseNet
ResponseNet v.3 is an enhanced version of ResponseNet, a webserver that is designed to highlight signaling and regulatory pathways connecting user-defined proteins and genes by using the ResponseNet network optimization approach (https://netbio.bgu.ac.il/respnet). Users run ResponseNet by defining source and target sets of proteins, genes and/or microRNAs, and by specifying a molecular interaction network (interactome). The […] More Info | Go to ResponseNet
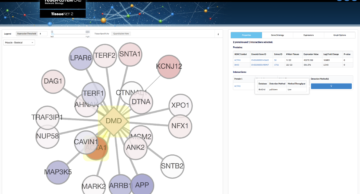
TissueNet
Knowledge of protein–protein interactions (PPIs) is important for identifying the functions of proteins and the processes they are involved in. Although data of human PPIs are easily accessible through several public databases, these databases do not specify the human tissues in which these PPIs take place. The TissueNet database of human tissue PPIs (https://netbio.bgu.ac.il/tissuenet/) associates […] More Info | Go to TissueNet
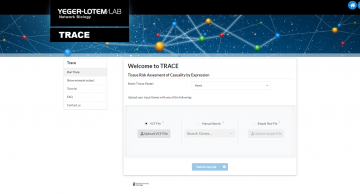
TRACE
‘Tissue Risk Assesment of Casuality by Expression’ accompanies the paper Simonovsky et al, Molecular Systems Biology 2023. TRACE is a catalog of the likelihood of protein-coding genes to lead to disease manifestation in eight human tissues.
More Info | Go to TRACE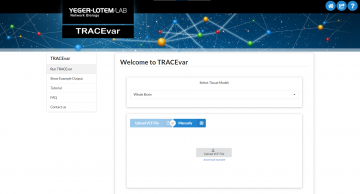
TRACEvar
Tissue Risk Assesment of Casuality by Expression for Variants’ accompanies the paper Argov et al., 2023. Given genetic variants and a specific tissue, TRACEvar estimates the likelihood that a given variant causes a hereditary disease that manifests clinically in the specified tissue.
More Info | Go to TRACEvar