What is ODiseA?
Organ-Disease Annotations (ODiseA) is a database that provides throroughly-curated annotations for hereditary diseases to their inflicted tissues.
Why use ODiseA?
Knowledge of the tissues inflicted by hereditary diseases can illuminate their underlying mechanisms, which often remain elusive (reviewed in Hekselman and Yeger-Lotem, Nat Rev Genet 2020). Owing to its focus on human diseases and its extensive, high-quality data, ODiseA provides a valuable and unique tool for computational biologists and geneticists studying hereditary diseases and their underlying mechanisms. Additional information can be found in studies by our lab (Barshir et al, PLoS Genet 2018; Hekselman and Yeger-Lotem, Nat Rev Genet 2020; Basha et al, Bioinformatics 2020 ; Shemesh et al, Nat Commun 2021).
What do you mean by “Inflicted” and “Pathogenic” tissues?
Disease-inflicted tissues are tissues that show main pathophysiological changes in diagnosed patients, based on clinical, laboratory, or imaging evidence. Pathogenic-tissues indicates whether the pathophysiological changes emerges in that tissue or in another tissue. For example, the disease ‘stress-induced myopathic carnitine palmitoyltransferase II deficiency’ (OMIM: 255110) is characterized by muscle breakdown, followed by renal failure due to renal myoglobin accumulation. Hence, both kidneys and skeletal muscle are inflicted, whereas only skeletal muscle is pathogenic.
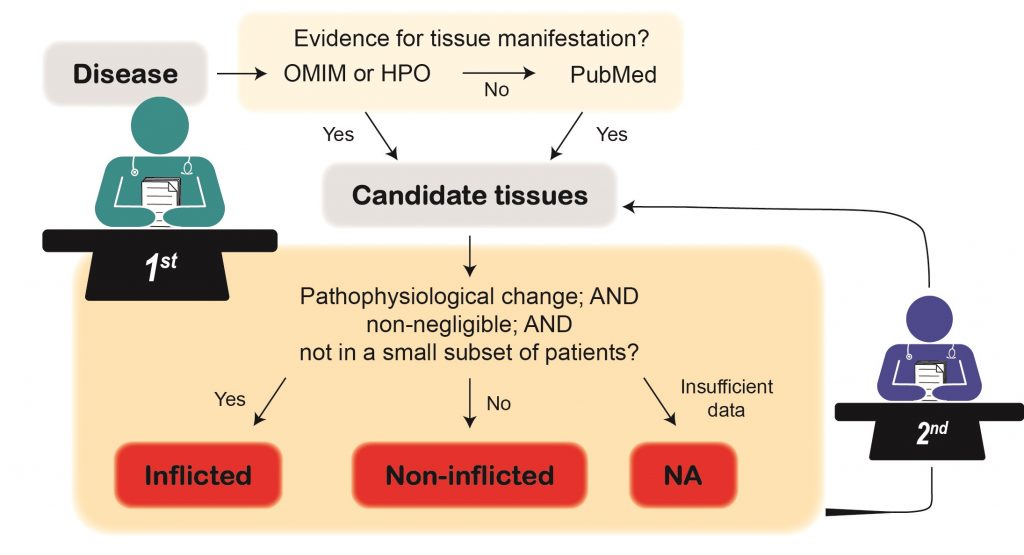
How were disease-inflicted tissues curated?
To curate disease-inflicted tissues, curators were randomly assigned a subset of diseases. For each disease, the curator searched for candidate inflicted tissues by querying OMIM and HPO, and in case of limited information also by querying PubMed. Next, a candidate tissue was annotated as inflicted if: (1) The evidence pointed to a pathophysiological change in that tissue; (2) The pathophysiological change was not negligible compared to disease-related changes in other tissues; and (3) The pathophysiological change was not documented in a small subset of patients. If any of these conditions were not met, the candidate tissue was annotated as non-inflicted. A candidate tissue that did not fit with the above was annotated as NA under the inflicted category. Eventually, all annotations were reassessed by a different curator based on the same criteria.
How many diseases and tissues are annotated?
ODiseA provides annotations for 2,181 diseases and their 1,920 causal genes, to 45 inflicted tissues. The numbers of diseases that inflict on each tissue are indicated in ODiseA’s main page for the full dataset.
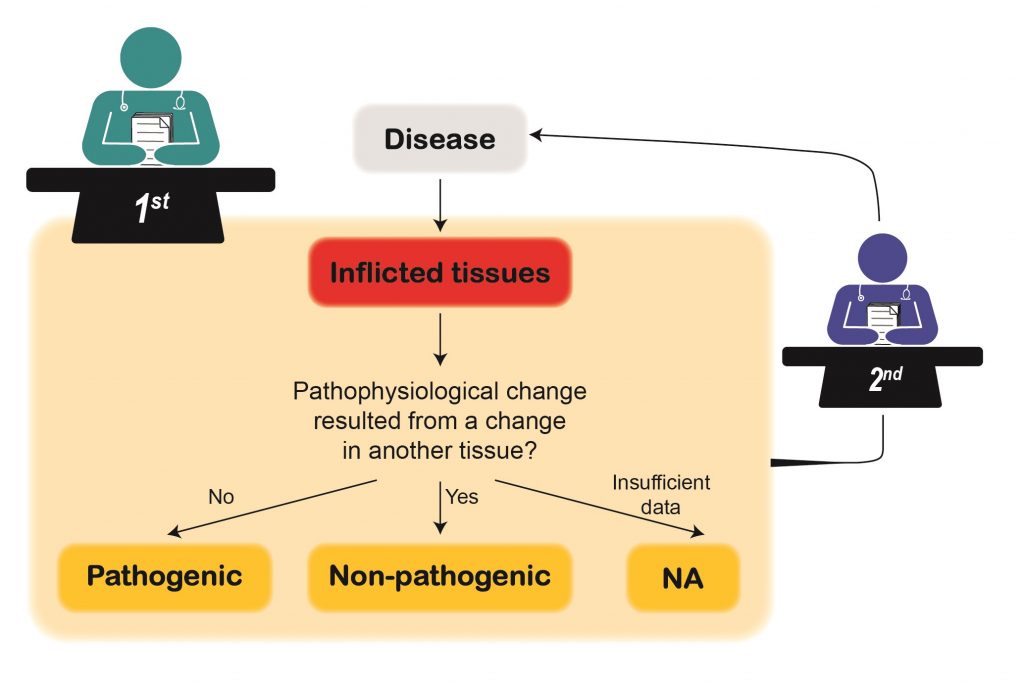
How were disease-pathogenic tissues curated?
To curate disease-pathogenic tissues curators annotated inflicted tissue as pathogenic if the pathophysiological change originated in that tissue. Otherwise, the tissue was annotated as non-pathogenic. Additionally, non-inflicted tissues were annotated as non-pathogenic. Eventually, all annotations were reassessed by a different curator based on the same criteria.
Where did the diseases extracted from?
Genetically-solved hereditary diseases were extracted from OMIM.
Why are some annotations empty?
Empty entries under the ‘Inflicted’ and ‘Pathogenic’ columns indicates that these annotations are yet to be assessed.
What is represented in the “Gene expression (TPM)” column?
The value under this column represents the expression level of the disease gene in the corresponding tissue. For this, transcriptomic profiles of non-diseased human tissues that matched tissues in ODiseA were downloaded from the GTEx portal version 8. Expression levels are measured in median transcripts per million [TPM; (GTEx Consortium, Science 2020)]. In case multiple tissues in GTEx corresponded to a single tissue in ODiseA (e.g., sun-exposed and non-exposed skin tissues in GTEx corresponded to skin tissue in ODiseA), the expression value of each gene was set to the maximal median TPM in the relevant tissues in GTEx.
What is represented in the “Gene ProAct” column?
This column links disease genes, which were annotated to biological processes by Gene Ontology, to Process Activity (ProAct) tool. ProAct estimates the activity of these processes in human tissues (Sharon et al, Bioinformatics 2022).
How to download the data?
All data can be downloaded from ODiseA’s main page > Download all data (to the left). Search output data can be downloaded from any search page > Download output data (at the top).

